Table of Contents
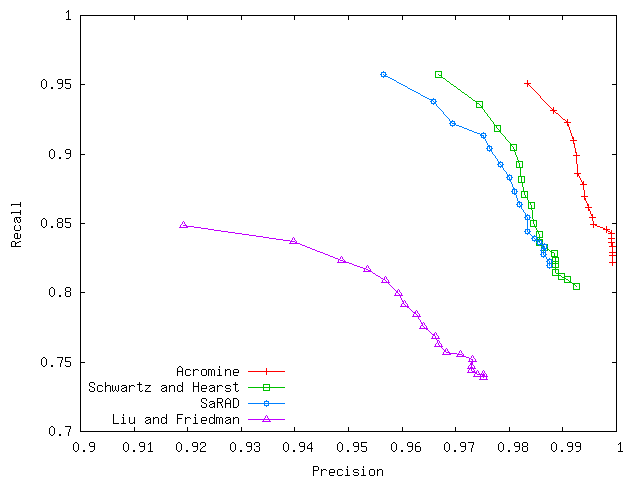
Acromine is a system for building a good quality acronym dictionary from running text. Assuming a word sequence co-occurring frequently with a parenthetical expression to be a potential expanded form, Acromine identifies acronym definitions in a similar manner to a statistical term recognition. Applied to the whole MEDLINE (7,811,582 abstracts) as of March 2006, Acromine extracted 920,425 acronym candidates and recognized 157,803 expanded forms in reasonable time (ca. 12 hours on a personal computer). This system achieves 99% precision and 82–95% recall on our evaluation corpus that roughly emulates the whole MEDLINE (Figure 1).
Please refer to the following paper for more detail.
-
[Okazaki06] “Building an abbreviation dictionary using a term recognition approach”. Bioinformatics. 2006. Oxford University Press.
You can try Acromine Acronym Dictionary at our demonstration site.
This section describes a tutorial to apply Acromine to the whole MEDLINE database.
The first step is to enumerate all short forms in a target text which are likely to be acronyms. All sentences containing a short form are inserted to an intermediate database for efficient access by later processes. Given a target text, we regard parenthetical expressions as short forms if all of the following conditions are met:
- they consist of at most two words
- their length is between two to ten characters
- they contain at least an alphabetic letter
- the first character is alphanumeric
To process MEDLINE XML files (*.xml.gz in this example) in the current directory, type the following command to construct a database acromine.shortform.db.
$ gzip -dc *.xml.gz | acromine_source_medline | acromine_shortform -c -d acromin e.shortform.db
The first process gzip decompress *.xml.gz files and sends their contents to the second process.
The second process acromine_source_medline sends the content of <AbstractText> and <ArticleTitle> elements to the third process.
The third process acromine_shortform recognizes short forms in the source text and stores contextual sentences of the short forms to a database.
Note that it takes time to process a huge amount of text. It took about 6 hours for Intel Core 2 Duo E6600 (2.40GHz, L2 4MB) processor with 2GB main memory to process the whole MEDLINE.
Once a shortform database is ready, acromine_shortform utility can retrieve all contextual sentences for a short form. For example, to retrieve all contextual sentences with the short form HMM, type the following command:
$ acromine_shortform --silent -d acromine.shortform.db -s HMM HMM 261 264 3512306:ABST:0_324 Limited proteolysis has been use d to study the influence of actin, in the absence or presence of regulatory prot eins of the thin filament (tropomyosin and troponin), as well as that of the myo fibrillar structure on the tryptic cleavage of the heavy meromyosin (HMM)/light meromyosin (LMM) hinge region in myosin heavy chain. HMM 263 266 10613897:ABST:0_307 The structural basis for the pho sphoryla- tion-dependent regulation of smooth muscle myosin ATPase activity was investigated by forming two- dimensional (2-D) crystalline arrays of expressed u nphosphorylated and thiophosphorylated smooth muscle heavy meromyosin (HMM) on p ositively charged lipid monolayers. HMM 13 16 2500343:ABST:355_455 Altretamine (HMM) (150 mg/m2) wa s administered orally days 2-8, therapy being resumed every 29 days. HMM 14 17 131797:ABST:0_68 H-Meromyosin (HMM) was digested with insoluble papain [EC 3.4.22.2]. HMM 271 274 6325466:ABST:0_300 Sixty-eight patients with "advan ced ovarian carcinoma" were entered into an ongoing phase-II trial for remission induction with cis-platinum (DDP) 80 mg/m2 i.v. on day 1 followed by forced sal ine diuresis, melphalan (L-PAM) 12 mg/m2 i.v. on day 2 and hexamethylmelamine (H MM) 130 mg/m2 p.o. HMM 18 21 236667:ABST:527_627 Heavy meromyosin (HMM) from cond itioned hearts had a higher Ca++-ATPase activity than from controls. ...
Each line in the output consists of five fields separated by tab characters:
- the target acronym,
- begin offset position, in bytes, of the acronym in the contextual sentence,
- end offset position, in bytes, of the acronym in the contextual sentence,
- PMID with begin/end offset positions, in bytes, of the contextual sentence in
<AbstractText>or<ArticleTitle>XML element, - the contextual sentence.
acromine_shortform utility also can enumerate all short forms stored in a database.
$ acromine_shortform -d acromine.shortform.db -l | sort -nr 54833 II 32921 CT 31294 III 27340 P<0.05 27016 PCR 24783 NO 20521 HIV 19154 LPS 19056 RA 18780 MRI 17721 P<0.001 17528 ELISA 17348 AD 16443 SD 16363 IV 15318 BP 14697 CSF 14691 MR 14642 P<0.01 14610 IL 14592 CNS 13557 PKC 13253 RT-PCR 13211 CONTROL ...
Note that sort command, which arranges lines in numerical order, is not a standard command on Windows environments but from Cygwin package. Each line in the output consists of two fields separated by a tab character:
- frequency of occurrence of a short form,
- the short form.
Finally, to extract long forms for a short form, run acromine_shortform to collect contextual sentences for the short form and acromine_longform to recognize long forms in the sentences. The following command extracts long forms for the short form HMM.
$ acromine_shortform --silent -d acromine.shortform.db -s HMM | acromine_longfor m HMM Acromine Longform Extractor version 1.0 Copyright (c) 2006 by N. Okazaki Shortform: HMM Candidates: 4326 entries generated. Scoring: done. HMM heavy meromyosin 238.983 245 HMM H-meromyosin 5 6 HMM hexamethylmelamine 52.9565 55 HMM hidden Markov model 113.547 116 HMM high molecular mass 27.9286 29 HMM human monocyte-macrophages 4 5 HMM human monocyte-derived macrophages 3 4 HMM human malignant mesothelioma 2 3 HMM hydroxymethylmexiletine 4.25 8
The utility acromine_longform outputs the recognition results to STDOUT. Each line of the output consists of four fields separated by tab characters:
- short form,
- long form,
- long form likelihood,
- frequency of occurrence of the short/long-form pair.
Iterating this process for all short forms yields a comprehensive acronym dictionary.
Acromine software utilizes following libraries:
- Oracle Berkeley DB is used for constructing a shortform database by acromine_shortform utility. It is licensed under Open Source License for Oracle Berkeley DB.
- zlib is used for compressing/decompressing contextual sentences by acromine_shortform utility. It is licensed under zlib License.
- Porter Stemmer is used for normalizing long forms by acromine_longform utility.
- optparse, strsplit, and quark are licensed under zlib License.